Login

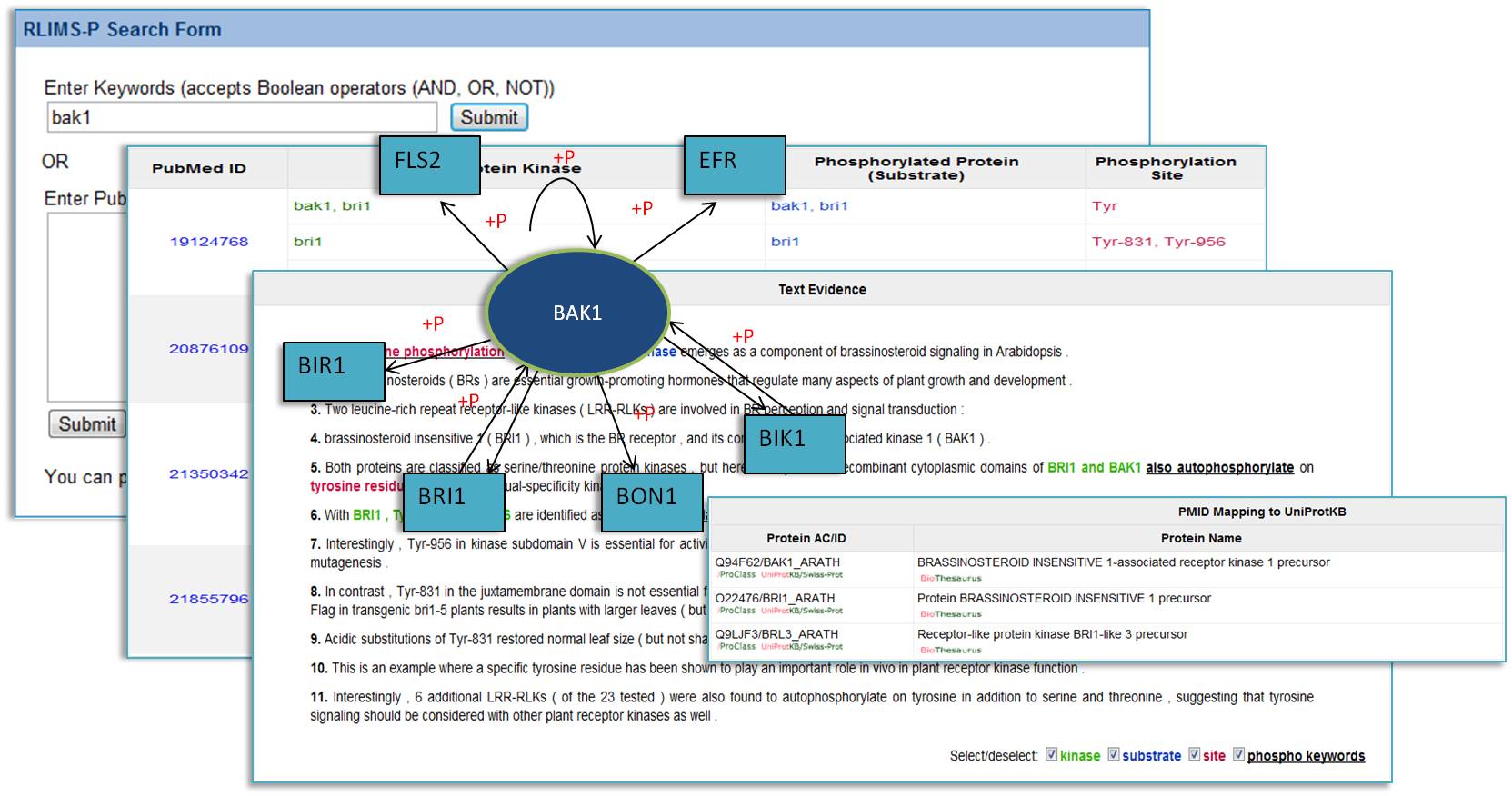
RLIMS-P is a rule-based text-mining program specifically designed to extract protein phosphorylation information on protein kinase, substrate and phosphorylation sites from biomedical literature (Hu et al., 2005). RLIMS-P currently works on PubMed abstracts, but it will be extended to open access full text articles soon.
RLIMS-P Search Form
Enter Keywords (accepts Boolean operators (AND, OR, NOT))
Or Enter PubMed IDs (PMIDs) delimited by "," or space, e.g., 15234272, 16436437.
You can process up to 200 PMIDs per run. Sample output
You can process up to 200 PMIDs per run. Sample output
This RLIMSP website enables user search phosphorylation information by keywords or a list of PMIDs in the database. The results (kinase, substrate, site) are displayed in sortable tables with text evidence and downloadable for further research.